Data Integration in Systems Biology: Characterization of Biological Phenomena from Structural and Functional Information
- 1418
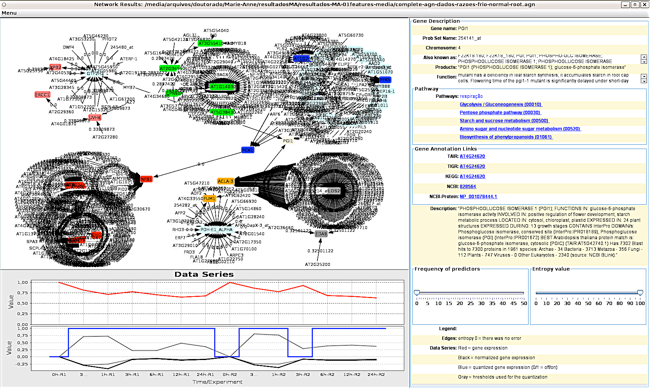
One of the most challenging research problem of System Biology nowadays is the inference (or reverse-engineering) of gene regulatory networks (GRNs) from expression profiles. This research issue became important after the development of high-throughput technologies for extraction of gene expressions, such as DNA microarrays or SAGE, and more recently RNA-Seq.
This problem regards on discover regulatory relationships between biological molecules in order to recover a complex network of interrelationships, which can reveal/describe not only diverse biological functions but also the dynamics of molecular activities. It is very important to understand how many biological processes happen and in most cases, how to prevent it from happening (diseases).
In the context of expression profiles, a big challenge that researchers need to face is the large number of variables or genes (thousands) for just a few experiments available (dozens). In order to infer relationships among those variables, it is needed a great effort in developing novel computational and statistical techniques that are able to alleviate the intrinsic error estimation committed in the presence of small number of samples with huge dimensionalities.
In general, it is not possible to recover the GRNs very accurately. The main reasons for this are the lack information about the biological organism, the high complexity of the networks and the intrinsic noise of the expression measurements. Thus, infer, analyze and compare the interrelationship between genes with precision, generating Gene Regulatory Networks (GRNs) is an open research problem.





